Recombinant West Nile virus Genome polyprotein, partial
-
中文名稱:Recombinant West Nile virus Genome polyprotein ,partial
-
貨號:CSB-YP356974WAF
-
規格:
-
來源:Yeast
-
其他:
-
中文名稱:Recombinant West Nile virus Genome polyprotein ,partial
-
貨號:CSB-EP356974WAF
-
規格:
-
來源:E.coli
-
其他:
-
中文名稱:Recombinant West Nile virus Genome polyprotein ,partial
-
貨號:CSB-EP356974WAF-B
-
規格:
-
來源:E.coli
-
共軛:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名稱:Recombinant West Nile virus Genome polyprotein ,partial
-
貨號:CSB-BP356974WAF
-
規格:
-
來源:Baculovirus
-
其他:
-
中文名稱:Recombinant West Nile virus Genome polyprotein ,partial
-
貨號:CSB-MP356974WAF
-
規格:
-
來源:Mammalian cell
-
其他:
產品詳情
-
純度:>85% (SDS-PAGE)
-
基因名:N/A
-
Uniprot No.:
-
別名:Genome polyprotein [Cleaved into: Peptide 2k; Capsid protein C; Core protein); Protein prM; Peptide pr; Small envelope protein M; Matrix protein); Envelope protein E; Non-structural protein 1; NS1); Non-structural protein 2A; NS2A); Serine protease subunit NS2B; Flavivirin protease NS2B regulatory subunit; Non-structural protein 2B); Serine protease NS3; EC 3.4.21.91; EC 3.6.1.15; EC 3.6.4.13; Flavivirin protease NS3 catalytic subunit; Non-structural protein 3); Non-structural protein 4A; NS4A); Non-structural protein 4B; NS4B); RNA-directed RNA polymerase NS5; EC 2.1.1.56; EC 2.1.1.57; EC 2.7.7.48; NS5)]
-
種屬:West Nile virus (WNV)
-
蛋白長度:Partial
-
蛋白標簽:Tag?type?will?be?determined?during?the?manufacturing?process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
產品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
復溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
儲存條件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保質期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
貨期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事項:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相關產品
靶點詳情
-
功能:Plays a role in virus budding by binding to the cell membrane and gathering the viral RNA into a nucleocapsid that forms the core of a mature virus particle. During virus entry, may induce genome penetration into the host cytoplasm after hemifusion induced by the surface proteins. Can migrate to the cell nucleus where it modulates host functions. Overcomes the anti-viral effects of host EXOC1 by sequestering and degrading the latter through the proteasome degradation pathway.; Inhibits RNA silencing by interfering with host Dicer.; Prevents premature fusion activity of envelope proteins in trans-Golgi by binding to envelope protein E at pH6.0. After virion release in extracellular space, gets dissociated from E dimers.; Acts as a chaperone for envelope protein E during intracellular virion assembly by masking and inactivating envelope protein E fusion peptide. prM is the only viral peptide matured by host furin in the trans-Golgi network probably to avoid catastrophic activation of the viral fusion activity in acidic Golgi compartment prior to virion release. prM-E cleavage is inefficient, and many virions are only partially matured. These uncleaved prM would play a role in immune evasion.; May play a role in virus budding. Exerts cytotoxic effects by activating a mitochondrial apoptotic pathway through M ectodomain. May display a viroporin activity.; Binds to host cell surface receptor and mediates fusion between viral and cellular membranes. Envelope protein is synthesized in the endoplasmic reticulum in the form of heterodimer with protein prM. They play a role in virion budding in the ER, and the newly formed immature particle is covered with 60 spikes composed of heterodimer between precursor prM and envelope protein E. The virion is transported to the Golgi apparatus where the low pH causes dissociation of PrM-E heterodimers and formation of E homodimers. prM-E cleavage is inefficient, and many virions are only partially matured. These uncleaved prM would play a role in immune evasion.; Involved in immune evasion, pathogenesis and viral replication. Once cleaved off the polyprotein, is targeted to three destinations: the viral replication cycle, the plasma membrane and the extracellular compartment. Essential for viral replication. Required for formation of the replication complex and recruitment of other non-structural proteins to the ER-derived membrane structures. Excreted as a hexameric lipoparticle that plays a role against host immune response. Antagonizing the complement function. Binds to the host macrophages and dendritic cells. Inhibits signal transduction originating from Toll-like receptor 3 (TLR3).; Component of the viral RNA replication complex that functions in virion assembly and antagonizes the host alpha/beta interferon antiviral response.; Required cofactor for the serine protease function of NS3. May have membrane-destabilizing activity and form viroporins.; Displays three enzymatic activities: serine protease, NTPase and RNA helicase. NS3 serine protease, in association with NS2B, performs its autocleavage and cleaves the polyprotein at dibasic sites in the cytoplasm: C-prM, NS2A-NS2B, NS2B-NS3, NS3-NS4A, NS4A-2K and NS4B-NS5. NS3 RNA helicase binds RNA and unwinds dsRNA in the 3' to 5' direction. NS3 supports the separation of RNA daughter and template strands during viral replication. The helicase part is involved in the inhibition of phosphorylation of host STAT1, and thereby inhibition of host type-I IFN signaling. In addition, NS3 assists the initiation of replication by unwinding the RNA secondary structure in the 3' non-translated region (NTR). Inhibits STAT2 translocation in the nucleus after IFN-alpha treatment.; Regulates the ATPase activity of the NS3 helicase activity. NS4A allows NS3 helicase to conserve energy during unwinding.; Functions as a signal peptide for NS4B and is required for the interferon antagonism activity of the latter.; Induces the formation of ER-derived membrane vesicles where the viral replication takes place. Inhibits interferon (IFN)-induced host STAT1 phosphorylation and nuclear translocation, thereby preventing the establishment of cellular antiviral state by blocking the IFN-alpha/beta pathway. Inhibits STAT2 translocation in the nucleus after IFN-alpha treatment.; Replicates the viral (+) and (-) RNA genome, and performs the capping of genomes in the cytoplasm. NS5 methylates viral RNA cap at guanine N-7 and ribose 2'-O positions. Besides its role in RNA genome replication, also prevents the establishment of cellular antiviral state by blocking the interferon-alpha/beta (IFN-alpha/beta) signaling pathway. Inhibits host TYK2 and STAT2 phosphorylation, thereby preventing activation of JAK-STAT signaling pathway.
-
基因功能參考文獻:
- evidence of the intrinsic flexibility and conformational heterogeneity of the NS2B chain of the WNV protease in the absence of substratelike ligands, which should be considered during antiviral drug discovery and development efforts PMID: 24015950
- Mass spectrometry and coimmunoprecipitation studies established a novel physical interaction between NS1 and NS4B, suggesting a mechanism for how luminal NS1 conveys signals to the cytoplasm to regulate RNA replication. PMID: 22553322
- NS4B P38G substitution was associated with both temperature-sensitive and small-plaque phenotypes. PMID: 22314017
- KAWGKSILFA is a novel epitope of NS1 protein recognized by the NS1 mAb. PMID: 21918007
- West Nile virus specific B-cell epitopes located at aa 21-36, 101-116, 191-206 and 261-276 of NS1. PMID: 21940411
- The combined effects of proline and negatively charged residues within the PEPE peptide are essential to promote the cleavage of 2K from NS4A, which is a prerequisite for efficient West Nile virus replication. PMID: 21880777
- Results highlights the importance of specific functional groups for the binding of the purine substrate, and reveals the complexity of the active site of the NS3 protein. PMID: 20421212
- The authors showed that the combination of the residues 156 and 159 of the E protein plays an important role in the transport of West Nile virus virus-like particles across endothelial cells. PMID: 20529314
- Dissociation of the C-terminal of the NS2B cofactor from the NS3 protease occurs in the presence and the absence of inhibitors. PMID: 19997625
- MKRN1 could induce WNV capsid protein ubiquitination and degradation in a proteasome-dependent manner. PMID: 19846531
- characterization and homology model of West Nile virus NS3 protease PMID: 15322074
- mutagenesis and kinetic studies of the West Nile Virus NS3 protease PMID: 15494419
- comparative studies revealed differential expression of epitopes within the E protein domain III of ten naturally occurring strains representing major subtypes of genetic lineages 1 and 2 PMID: 15823609
- NS3 alone triggers the apoptotic pathways involving caspases-8 and -3 PMID: 16243374
- A30P mutation results in virus which elicits more rapid induction & higher levels of synthesis of IFN-alpha/beta in infected cells than wild-type; results confirm & extend previous findings on role of NS2A in inhibition of host antiviral response PMID: 16474146
- cleavage of the Australian strain of West Nile virus, Kunjin virus (KUNV), polyprotein NS4A-4B by the viral protease is the key initiation event in the induction of membrane rearrangement PMID: 16611922
- the C102S mutation of NS4B was associated with a temperature-sensitive phenotype at 41 degrees C as well as attenuation of the neuroinvasive and neurovirulence phenotypes in mice. PMID: 16624366
- NS5 sequentially catalyzes guanine-N-7 and ribose 2'-O methylations invovled in RNA cap formation [NS5] PMID: 16912287
- crystal structure of a soluble fragment of West Nile virus E; structure adopts the same overall fold as that of E proteins from dengue and tick-borne encephalitis viruses; conformation of domain II is different from that in other prefusion E structures PMID: 16943291
- results disclosed the strict substrate specificity of the WNV NS2B-NS3 proteases for which the (K/R)(K/R)R/GG amino acid motifs was optimal PMID: 17067286
- established for the first time that the RNA-dependent RNA polymerase NS5 is located in flavivirus-induced membranes, including the site of viral RNA replication PMID: 17374759
- WNV-mediated neuroinflammation and cell death occurred through WNV infection of both glia and neurons, which was driven in part by WNV capsid protein expression PMID: 17670819
- WNV envelope protein (WNV-E) specifically blocks the production of antiviral and proinflammatory cytokines induced by dsRNA in murine macrophages. PMID: 18056386
- Molecular modelling studies suggest a difference in local structure of the E protein associated with either an asparagine or serine residue at position 155 compared with the tyrosine found in the virulent parental WN-Israel virus. [E protein] PMID: 18272752
- Two novel NS2B functional determinants critical for NS3pro activation were identified. [NS2B] PMID: 18272753
- The results further confirm the role of NS2A in virus assembly, demonstrate the importance of hydrophobic residues at codon 59 in this process, implicate the involvement of NS2A in the biogenesis of virus-induced membranes PMID: 18337583
- analysis of the two-component NS2B-NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase PMID: 18442976
- The linker region plays a critical role in determining the NS3 oligomerization that leads to the formation of the active ATPase/helicase oligomer. PMID: 18448185
- West Nile virus NS5 binds specifically to the 5'-terminal stem-loop (SL1) of the genomic RNA and this structure is essential for WNV replication. PMID: 18799181
- data demonstrate that an intact West Nile virus fusion loop peptide of the envelope protein is critical for virulence, and that human mAb11 targeting this region is efficacious against West Nile virus infection PMID: 19535627
- These data support a structural model of the full-length NS5 molecule that predicts a physical interaction between the MTase and the RdRp domains. [[NS5] PMID: 19710254
- These results do not support a requirement for any single histidine as a pH-sensing "switch," and they suggest that additional features of the E protein are involved in triggering pH-dependent steps in the flavivirus life cycle. PMID: 19776132
顯示更多
收起更多
-
亞細胞定位:[Capsid protein C]: Virion. Host nucleus. Host cytoplasm. Host cytoplasm, host perinuclear region.; [Peptide pr]: Secreted.; [Small envelope protein M]: Virion membrane; Multi-pass membrane protein. Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Envelope protein E]: Virion membrane; Multi-pass membrane protein. Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Non-structural protein 1]: Secreted. Host endoplasmic reticulum membrane; Peripheral membrane protein; Lumenal side.; [Non-structural protein 2A]: Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Serine protease subunit NS2B]: Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Serine protease/Helicase NS3]: Host endoplasmic reticulum membrane; Peripheral membrane protein; Cytoplasmic side.; [Non-structural protein 4A]: Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Non-structural protein 4B]: Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [RNA-directed RNA polymerase NS5]: Host endoplasmic reticulum membrane; Peripheral membrane protein; Cytoplasmic side. Host nucleus. Host cytoplasm.
-
蛋白家族:Class I-like SAM-binding methyltransferase superfamily, mRNA cap 0-1 NS5-type methyltransferase family
-
數據庫鏈接:
KEGG: vg:912267
Most popular with customers
-
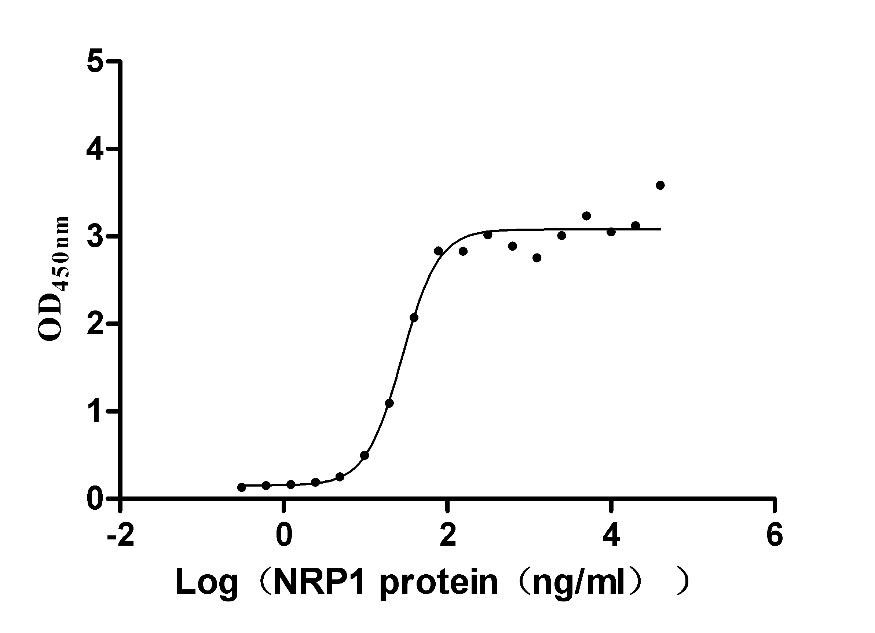
Recombinant Human Neuropilin-1 (NRP1) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
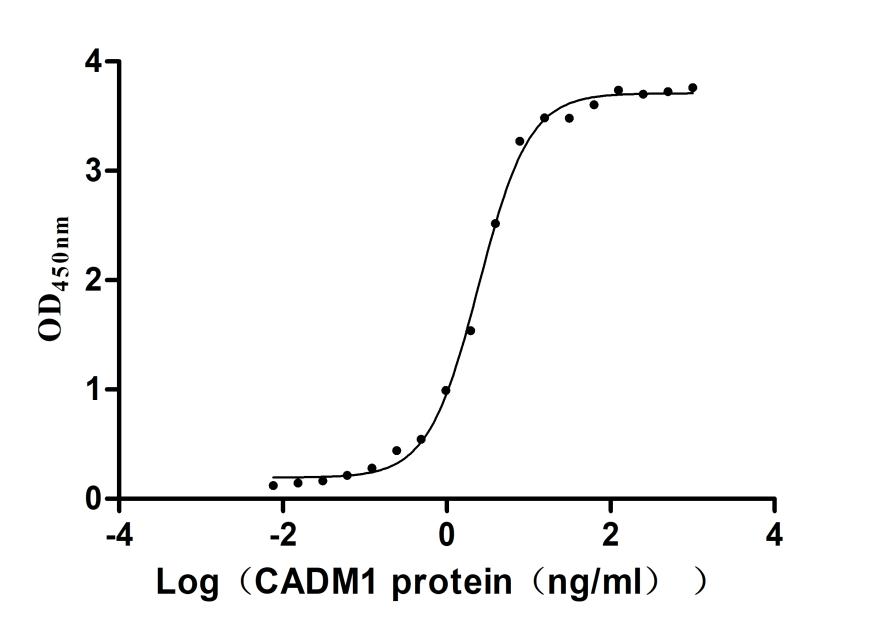
Recombinant Human Nectin-4 (NECTIN4), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Cytotoxic and regulatory T-cell molecule (CRTAM), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
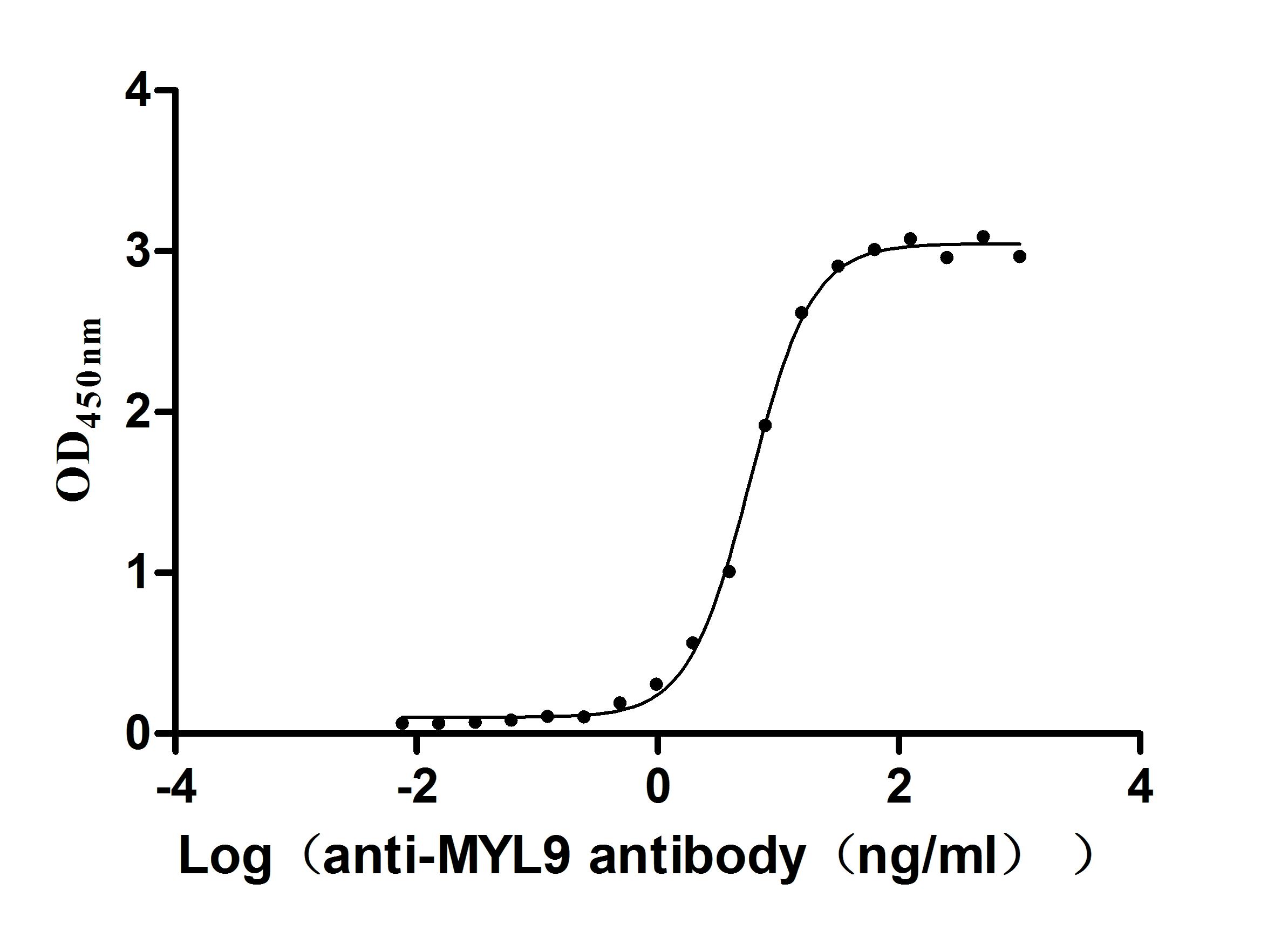
Recombinant Human Myosin regulatory light chain 12A (MYL12A) (Active)
Express system: E.coli
Species: Homo sapiens (Human)